Article Text
Abstract
Background Epigenetic alterations accumulate in normal-appearing tissues of patients with cancer, producing an epigenetic field defect. Cross-sectional studies show that the degree of the defect may be associated with risk in some types of cancer, especially cancers associated with chronic inflammation.
Objective To demonstrate, by a multicentre prospective cohort study, that the risk of metachronous gastric cancer after endoscopic resection (ER) can be predicted by assessment of the epigenetic field defect using methylation levels.
Design Patients with early gastric cancer, aged 40–80 years, who planned to have, or had undergone, ER, were enrolled at least 6 months after Helicobacter pylori infection discontinued. Methylation levels of three preselected genes (miR-124a-3, EMX1 and NKX6-1) were measured by quantitative methylation-specific PCR. Patients were followed up annually by endoscopy, and the primary endpoint was defined as detection of a metachronous gastric cancer. Authentic metachronous gastric cancers were defined as cancers excluding those detected within 1 year after the enrolment.
Results Among 826 patients enrolled, 782 patients had at least one follow-up, with a median follow-up of 2.97 years. Authentic metachronous gastric cancers developed in 66 patients: 29, 16 and 21 patients at 1–2, 2–3 and ≥3 years after the enrolment, respectively. The highest quartile of the miR-124a-3 methylation level had a significant univariate HR (95% CI) (2.17 (1.07 to 4.41); p=0.032) and a multivariate-adjusted HR (2.30 (1.03 to 5.10); p=0.042) of developing authentic metachronous gastric cancers. Similar trends were seen for EMX1 and NKX6-1.
Conclusions Assessment of the degree of an epigenetic field defect is a promising cancer risk marker that takes account of life history.
- GASTRIC CANCER
- METHYLATION
This is an Open Access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 3.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/3.0/
Statistics from Altmetric.com
Significance of this study
What is already known about this subject?
-
Epigenetic alterations, represented by aberrant DNA methylation, accumulate in cancers and also in normal-appearing tissues surrounding cancers.
-
Cross-sectional studies show that aberrant methylation levels in normal tissues may be associated with a risk of cancer in some types of cancer, especially chronic inflammation-associated cancers.
-
An epigenetic cancer risk marker is considered to reflect past exposure to environmental factors and to differ from single nucleotide polymorphism cancer risk markers that do not reflect life history.
What are the new findings?
-
By a multicentre prospective cohort study with 826 patients, the methylation level in non-cancerous gastric mucosae of patients with gastric cancer was shown to be significantly (p=0.042) associated with an increased risk of developing metachronous gastric cancers.
-
miR-124a-3 is an informative marker gene for predicting the risk of developing a metachronous gastric cancer.
How might it impact on clinical practice in the foreseeable future?
-
This is the first time that the usefulness of an epigenetic cancer risk marker has been demonstrated in any type of cancer by a multicentre prospective cohort study. This new class of cancer risk marker has the potential to be expanded to cancers of other tissues.
-
The intensity of surveillance for metachronous gastric cancer can be adjusted depending upon the risk predicted by the miR-124a-3 methylation level.
Introduction
Epigenetic alterations, represented by aberrant DNA methylation, are deeply involved in carcinogenesis.1 ,2 Aberrant methylation accumulates in cancers and also in normal-appearing tissues, especially those of chronic inflammation-associated cancers, such as gastric cancers,3 ,4 hepatocellular carcinoma,5 ,6 oesophageal adenocarcinoma,7 ,8 colon cancers,9 colitic cancers10–12 and breast cancers.13 In such normal-appearing tissues, both tumour-suppressor genes (driver genes), such as CDKN2A and MLH1 and many other genes that have little expression in normal tissues (passenger genes), such as HAND1, are methylated. Driver genes are methylated only at low levels—that is, in a minor fraction of cells, in normal-appearing tissues. Passenger genes, however, are methylated at high levels—that is, in a large fraction of cells, reflecting the degree of past exposure to inducers of aberrant methylation.3 ,14
The accumulation of such methylation was shown to be associated with a risk of cancer development by cross-sectional studies in the stomach,14 the liver,5 ,6 the urothelium,15 the oesophagus8 ,16 and the colon.17 In the stomach, a quantitative methylation analysis of both driver and passenger genes showed that patients with gastric cancer had higher methylation levels in normal-appearing tissues than those in healthy individuals.14 ,18 Furthermore, patients with multiple gastric cancers had significantly higher methylation levels than those with a single gastric cancer, whose methylation levels were higher than those in healthy individuals.19 This correlation in the three groups, together with the associations in various types of cancers, strongly supports the suggestion that the accumulation of aberrant methylation in non-cancerous tissues produces an epigenetic field for cancerisation (field defect) and that the presence of such a field is associated with cancer development.3
As an inducer of an epigenetic field defect in the stomach, Helicobacter pylori (H. pylori) infection was associated with high methylation levels in gastric mucosae.14 Chronic inflammation triggered by the infection was shown to be causally involved in methylation induction in Mongolian gerbils.20 It is hypothesised that aberrant DNA methylation is induced in stem, progenitor and differentiated cells when active H. pylori infection is present3 because eradication of H. pylori leads to a decrease of methylation.21–23 Importantly, the methylation levels in gastric mucosae without active H. pylori infection, but not those in mucosae with active infection, were correlated with gastric cancer risk.14 ,19 ,24 Therefore, measurement of methylation levels in individuals without active H. pylori infection is expected to enable us to predict the cancer risk of an individual by analysing the degree of epigenetic field defect.
For incorporation of such a new type of cancer risk marker into practice, demonstration of its usefulness by a multicentre prospective cohort study is requisite. In gastric cancer, a prospective cohort study can be conducted for risk prediction of primary gastric cancer among healthy individuals and for prediction of a metachronous gastric cancer after endoscopic resection (ER). The need for the latter is becoming greater because an increasing number of patients with gastric cancer are now treated by ER and the incidence of metachronous gastric cancer has reached as high as 2.5% a year,25 although eradication of H. pylori decreases or delays its occurrence.26 Nevertheless, almost all patients treated by ER have similar risk factors and their stratification for future cancer risk has so far been impossible. In addition, this high incidence allows a prospective cohort study with a relatively small number of patients.
Here, we aimed to demonstrate, by a multicentre prospective cohort study, that the risk of metachronous gastric cancer can be predicted by assessment of an epigenetic field defect. This will provide a proof-of-concept that cancer risk prediction can be achieved using the epigenetic field defect.
Patients and methods
Patients
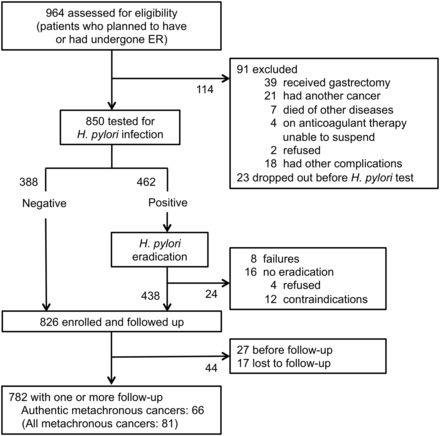
Eligibility was assessed for 964 patients with early gastric cancer aged 40–80 years and who planned to have, or had undergone, endoscopic submucosal dissection, one of the procedures of ER, in one of three hospitals (National Cancer Center Hospital (NCC), Tokyo University Hospital (TYU) and Wakayama Medical University Hospital (WMU)) between 2008 and 2010. Patients were excluded if they had received additional gastrectomy, had cancer in other tissues, died of other diseases before a test for H. pylori infection or were receiving anticoagulant therapy and unable to suspend it, refused or had other complications. The remaining 850 patients were tested for H. pylori infection. When H. pylori infection was absent, they were enrolled at the assessment (n=388). When H. pylori infection was present (n=462), patients received eradication therapy and were enrolled at least 6 months after establishment of successful H. pylori eradication (n=438) (figure 1). A total of 826 patients were enrolled and at the time of enrolment, biopsy samples were taken from a fixed point in the antrum (the lesser curvature at 2 cm from the pyloric ring) for DNA methylation analysis. Written informed consent was obtained from all the patients and the study was approved by the institutional review boards at each hospital.
Study profile. ER, endoscopic resection.
Detection of H. pylori infection, its eradication and the pepsinogen test
The presence of H. pylori infection was examined with the urea breath test (Otsuka, Tokushima, Japan), serum anti-H. pylori antibody (Eiken, Tokyo, Japan) and either the culture method or rapid urease test (Otsuka, Tokushima, Japan). To eradicate H. pylori, patients were treated with lansoprazole (30 mg), amoxicillin (750 mg) and clarithromycin (200 mg), each taken twice daily for 1 week. Successful eradication was established by negative results of the urea breath test 2 months or more after the eradication. When the eradication was unsuccessful, the patients received second-line eradication therapy. Implementation of third-line and further eradication therapy was left to the doctors in charge. Fasting blood samples were collected on the day of endoscopy at the enrolment and serum levels of pepsinogen I and II were measured by the LZ test (Eiken, Tokyo, Japan).
Follow-up by endoscopy and definition of metachronous cancers
Patients were followed up endoscopically once a year after the enrolment and the primary endpoint was defined as detection of a metachronous gastric cancer. If a patient was lost to follow-up, the follow-up was censored at the time of their last endoscopic examination. Metachronous cancers were defined according to the criteria of Moertel et al27—namely, (i) each lesion is histopathologically malignant, (ii) each lesion is separated from another and (iii) each lesion is not the result of a local extension or metastasis of other lesions. In particular, ‘authentic’ metachronous gastric cancers were defined as those found after exclusion of cancers that developed within the first 1 year after the enrolment. This was because there is concern that a gastric cancer developing within the first 1 year after enrolment might have been a cancer undetected at the time of the emrolment28–30 and might be influenced by the promoting effect of H. pylori infection.26
Marker genes and quantitative methylation analysis
Three marker genes (miR-124a-3, EMX1 and NKX6-1) were preselected in our previous cross-sectional studies,31 ,32 and no other marker genes were analysed to avoid multiple testing. miR-124a-3 was previously identified as a gene whose methylation levels remain high, even in individuals with past H. pylori infection.31 EMX1 and NKX6-1 were identified as genes highly informative for detecting patients with gastric cancer among patients with past H. pylori infection with ORs of 23.8 and 15.0, respectively.32 The methylation levels of the three genes were analysed by quantitative methylation-specific PCR (qMSP), as previously described.31 ,32 Briefly, bisulfite modification was performed using 1 µg of BamHI-digested genomic DNA. qMSP was performed with primer sets specific to methylated and unmethylated sequences by real-time PCR using SYBR Green I (BioWhittaker Molecular Applications, Rockland, Maine, USA) and an iCycler thermal cycler (Bio-Rad Laboratories, Hercules, California, USA). To correct for the variation in the number of methylated and unmethylated DNA molecules, depending upon the dilution of the standard DNA, two specific batches of control DNA (fully methylated and unmethylated DNA) were analysed in each qMSP analysis. All the samples were measured twice and the correlation of methylation levels in the two analyses was high (correlation coefficient=0.94).
Statistical analysis
Our study question was whether, at any time, the methylation level in gastric mucosae could predict the risk of gastric cancer and we counted person-years since the enrolment (ie, since the time of biopsy of gastric mucosa).
To analyse the relationship between the methylation levels and a risk of metachronous gastric cancer, all the patients were categorised into quartiles (Q) according to the methylation level of one of the three genes (miR-124a-3, EMX1 and NKX6-1), with the baseline characteristics of Q1 (lowest) and Q4 (highest) shown in table 1. Since we assumed that a higher methylation level would be associated with a risk of metachronous gastric cancer, we estimated the HR and 95% CI of Q4 compared with Q1 as the major goal.
Baseline characteristics of patients according to the quartiles of DNA methylation levels of the three genes
Correlations between methylation levels of two genes were analysed using Spearman's rank correlation coefficient. Cox proportional hazard regression analysis was used to calculate univariate and multivariate-adjusted HRs (95% CIs) of metachronous gastric cancer incidence. Kaplan–Meier analysis was performed to compare development of metachronous gastric cancer in the four quartiles. The proportional hazard assumptions for the methylation level in comparing Q4 with Q1 were evaluated and met using graphical and time-dependent variable approaches. Analyses were conducted using the SAS statistical software, V.9.1 (SAS Institute Inc, Cary, North Carolina, USA). The results were considered significant when a p value <0.05 was obtained by two-sided tests.
In multivariate analysis, HRs were adjusted for potentially confounding factors (age (<50, 50–59, 60–69 or ≥70 years), gender, pepsinogen index ((−), pepsinogen I>70 or pepsinogen I/II ratio >3; (+), pepsinogen I≤70 and pepsinogen I/II ratio ≤3; (2+), pepsinogen I≤50 and pepsinogen I/II ratio ≤3; (3+), pepsinogen I≤30 and pepsinogen I/II ratio ≤2), smoking (0, 1–39 or ≥40 pack-years), green vegetable intake (≤2 days, 3–4 days or almost daily), hospital (NCC, TYU or WMU), H. pylori infection before the enrolment (positive or negative) and past history of ER (0, 1, 2 or 3 times)).
Results
Enrolment and follow-up for a metachronous gastric cancer
Among the 826 enrolled patients, 782 patients (617, 97 and 68 patients at NCC, TYU and WMU, respectively) had at least one follow-up endoscopic examination (figure 1). Among the 782 patients, 81 patients (59, 10 and 12 patients in NCC, TYU and WMU, respectively) developed a metachronous gastric cancer. Among the 81 patients, 15, 29, 16 and 21 patients developed a metachronous cancer in <1, 1–2, 2–3 and ≥3 years after the enrolment, respectively, and 66 patients (52, 6 and 8 patients in NCC, TYU and WMU, respectively) were considered to have authentic metachronous gastric cancers. The median follow-up period was 2.97 years (2.99, 2.51 and 2.49 years in NCC, TYU and WMU, respectively). The median period for development of all the metachronous cancers after the enrolment was 1.88 years (1.92, 1.97 and 1.32 years in NCC, TYU and WMU, respectively).
Univariate analysis of the effect of methylation levels
The methylation levels of miR-124a-3 and EMX1 showed bimodal distribution while that of NKX6-1 showed a unimodal distribution (figure 2). The methylation levels of the three genes were highly correlated (correlation coefficients=0.70–0.89; table 2), suggesting that methylation of these three genes reflected a shared mechanism, the epigenomic damage in gastric stem cells.
Correlation among methylation levels of miR-124a-3, EMX1, and NKX6-1
The distribution of methylation levels of miR-124a-3, EMX1 and NKX6-1 among the 782 patients. miR-124a-3 and EMX1 showed bimodal distributions, whereas NKX6-1 showed a unimodal distribution.
A univariate analysis using the authentic metachronous gastric cancers showed that Q4 (highest) miR-124a-3 methylation had a significantly higher HR than Q1 (lowest) (95% CI) (2.17 (1.07 to 4.41); p=0.032) (p for trend=0.041) (table 3). The presence of the same trend in Q2 and Q3 with a trend p of 0.041 supported the higher HR in the Q4. Although not significant, similar trends were seen for EMX1 and NKX6-1 (p=0.075 and 0.11, respectively). A univariate analysis using all the metachronous gastric cancers also showed that Q4 miR-124a-3 methylation had a higher HR than Q1 (95% CI) (1.71 (0.90 to 3.22); p=0.10) (p for trend=0.094) (table 3).
Univariate HRs (95% CI) of quartiles of the three genes for a metachronous gastric cancer
The influence of known risk factors for gastric cancer (age, gender, pepsinogen index, smoking and green vegetable intake) and other potential risk factors (hospital, H. pylori infection before enrolment, past history of ER) was analysed using the authentic metachronous gastric cancers. Significant associations were seen for young age (HR (95% CI)=3.50 (1.23 to 10.0); p=0.019), being female (0.43 (0.19 to 1.00); p=0.049), past history of ER (5.66 (2.01 to 15.9); p=0.001) and smoking (2.24 (1.27 to 3.96); p=0.006) (table 4). For all the metachronous gastric cancers, in addition to these factors, different hospitals had significantly different HRs (2.26 (1.21 to 4.21); p=0.01).
Univariate HRs (95% CI) of known and potential risk factors for metachronous gastric cancer
Multivariate analysis and Kaplan–Meier analysis
A multivariate analysis was conducted by adjusting for hospital, gender, age, H. pylori infection before enrolment, pepsinogen index, past history of ER, smoking and green vegetable intake (table 5). Using the authentic metachronous gastric cancers, the Q4 (highest) miR-124a-3 methylation had a higher HR than Q1 methylation (95% CI) (2.30 (1.03 to 5.10); p=0.042) (p for trend=0.057). when all the metachronous gastric cancers were used, Q4 miR-124a-3 methylation also had a higher HR (95%CI) (1.99 (0.97 to 4.09); p=0.061) (p for trend=0.072). We also conducted a multivariate analysis using the HR of the Q4 with the combined Q1–Q3 as a reference (see online supplementary table S1) and confirmed an association between the high miR-124a-3 methylation level and the occurrence of authentic metachronous gastric cancers (HR (95% CI)=1.95 (1.11 to 3.43; p=0.021)).
Multivariate-adjusted HRs (95% CI) for a metachronous gastric cancer according to DNA methylation levels of the three genes
Kaplan–Meier curves of the cumulative incidence rates of all the metachronous gastric cancers for patients were drawn for quartiles (Q1–Q4) of methylation levels for each of the three genes (miR-124a-3, EMX1 and NKX6-1) (figure 3 and see online supplementary figure S1). Q4 methylation had higher incidences of a metachronous gastric cancer for all the three genes compared with Q1 methylation.
Cumulative incidence of all the metachronous gastric cancers for patients in quartile (Q1–Q4) of methylation levels of miR-124a-3, EMX1, and NKX6-1. The Q4 methylation had higher incidences of a metachronous gastric cancer than the Q1 methylation with p values of 0.17, 0.08 and 0.54 for miR-124a-3, EMX1 and NKX6-1, respectively, by the log-rank test.
Analysis of potential confounding factors
The influence of H. pylori infection status before the enrolment did not affect the occurrence of metachronous gastric cancers (table 4). However, taking its potential influence on the predictive power of the methylation level, we conducted a stratified analysis by the H. pylori infection status before the enrolment. The median methylation levels of the three marker genes were lower in patients without H. pylori infection than those in patients with H. pylori infection (see online supplementary table S2). In the stratified analysis, the Q4 (highest) miR-124a-3 methylation levels showed HRs (95% CI) of 1.32 (0.37 to 4.77) and 2.43 (0.81 to 7.32) for the authentic metachronous gastric cancers in the patients with and without H. pylori infection, respectively (see online supplementary table S3).
The influence of a past history of ER on the predictive power of the methylation level was also analysed by stratifying patients according to this past history. Again, we observed consistent results in each stratum, although the p values were not statistically significant owing to the decreased numbers of events (see online supplementary table S4). Also, we analysed the variability in the time between the first ER and enrolment in each quartile (see online supplementary table S5). There were no increasing or decreasing trends in the time according to the marker quartiles. Additionally, we conducted a stratified analysis according to the time between the first ER and enrolment (>5 or ≤5 years) and observed a similar association in each stratum (see online supplementary table S4).
Discussion
It was shown for the first time by a multicentre prospective cohort study that assessment of an epigenetic field defect using methylation levels can be used as a biomarker of cancer risk. This study warrants translation of previous retrospective cross-sectional studies showing that methylation levels accumulated in various tissues are correlated with cancer risk.6 ,16 ,17 ,19 ,33 ,34 Thus, the intensity of surveillance for metachronous gastric cancer can be adjusted according to the risk predicted by the miR-124a-3 methylation level. In addition, methylation accumulation is known to be caused by exposure to its inducers, such as chronic inflammation,35 and cancer risk prediction using accumulated methylation is considered to take account of the life history of individuals. This point makes epigenetic cancer risk markers entirely different from genetic cancer risk markers, mostly single nucleotide polymorphisms, which cannot take account of the life history of individuals.
We defined the authentic metachronous gastric cancers because distinction of a metachronous cancer from that undetected at the time of the enrolment is difficult when it is detected at 1 year after the enrolment.28–30 Also, metachronous gastric cancer developing within 1 year after enrolment might have been influenced by the promoting effect of H. pylori infection.26 Analyses using both the authentic and all the metachronous gastric cancers were conducted and a clearer difference was seen using the authentic metachronous gastric cancers. For example, the methylation level of miR-124a-3 was associated with development of an authentic metachronous gastric cancer with significant p values of 0.032 and 0.042 by univariate and multivariate analyses, respectively. This supported the hypothesis that the presence of an epigenetic field defect is associated with occurrence of independent multiple cancers.
The quality of this translational study was supported by multiple parameters. The follow-up rate of patients enrolled in this study was high (97.9%; 809/826). The median follow-up period reached 2.97 years and was uniform in the three hospitals. Accordingly, the bias introduced by incomplete follow-up is minimised. Also, all the patients were followed up with a consistent interval of 1 year in the three participating hospitals. This is reflected in the Kaplan–Meier curves, which show that the incidence of a metachronous gastric cancer increases every 1 year. Further, even after adjusting for multiple variables, the influence of methylation levels on the occurrence of authentic metachronous gastric cancer was significant. The fact that two variables (gender and smoking) known to be associated with gastric cancer risk36 were confirmed as independent risk factors supported the statement that collection of lifestyle information was appropriately conducted in this study.
Nevertheless, several limitations of the study should be mentioned. First, the patient population was heterogeneous at enrolment for the past history of ER or the years since the first ER. These factors were associated with the occurrence of metachronous gastric cancer (table 4). Therefore, in addition to the multivariate analysis involving these factors, we conducted analyses stratified by these factors and found consistent results in each stratum, although the results were not statistically significant owing to the limited number of events (see online supplementary table S4). Thus, it is unlikely that these factors biased the association between the quartiles of miR-124a-3 and metachronous gastric cancer. Second, owing to a relatively small number of events, inevitable for a prospective study of cancer risk, we could not conclude which pattern, dose–response or threshold pattern, was obeyed by the relationship between methylation level of miR-124a-3 and cancer risk. However, we found an association of miR-124a-3 with authentic gastric cancer risk for the Q4 (highest) using Q1 (lowest) or the combination of all the other quartiles (Q1–Q3) as a reference (table 5 and see online supplementary table S1). This finding may support the threshold curve. Since the incidence of a metachronous gastric cancer is stable long after H. pylori eradication,25 ,37 further long-term follow-up is expected to strengthen the correlation and clarify an appropriate model of relationship.
From a molecular viewpoint, we tried to measure the accumulation of aberrant methylation in stem cells by analysing gastric mucosae without H. pylori infection, at least 6 months after H. pylori eradication. It is known that the methylation level in gastric mucosa decreases after H. pylori eradication,21–23 and that the persistent methylation level in gastric mucosa without H. pylori infection is correlated with gastric cancer risk.14 ,19 ,24 Since methylation induced in stem cells can only persist for a long time without its inducer, the methylation level in gastric mucosa without H. pylori infection could be considered to reflect epigenomic damage accumulated in stem cells. However, we do not have direct evidence to support the hypothesis, because it is still impossible to analyse DNA methylation of specific genes in histological sections. The higher methylation level and smaller HR in patients with H. pylori infection before the enrolment (see online supplementary table S3) might have indicated that the methylation levels had not reached baseline after eradication and were superimposed with methylation in progenitor cells.
We analysed three preselected marker genes. Ideal marker genes should be methylated in parallel with overall methylation levels of driver genes in stem cells, but at much higher levels for accurate measurement. miR-124a-3 is a tumour-suppressor gene (driver gene) with relatively high methylation levels,31 and was considered to have met the criteria. Two other genes (EMX1 and NKX6-1) were homeobox genes and considered to be passenger genes. In addition to the analysis using the methylation level of a single gene, we performed an exploratory analysis by combining methylation of three marker genes. Simple addition of the methylation levels of the three genes did not improve the prediction power (data not shown). However, when we scored the number of genes within the Q4 (highest) using the three marker genes, the patients with the highest scores displayed a high HR (95% CI) (2.23 (1.12 to 4.44); p=0.022) with a statistically significant trend p of 0.038 using the authentic metachronous gastric cancers. The smaller p value in this combined analysis also supports a threshold model.
In our previous study, we performed biopsies from three positions (antrum, middle body and upper body).19 It was shown that methylation levels of the three positions were different and that their mean had the highest association with gastric cancer risk. However, patients with a high mean methylation level had higher methylation levels in any of the three positions. In the current study, we performed biopsy from one position, considering the merits (smaller risk of bleeding) and demerits (less precise reflection of the cancer risk) of limiting the positions of biopsy. Since the utility of epigenetic analysis of gastric tissue was demonstrated here, an increase of biopsy positions in future studies may be considered.
In summary, the methylation level of miR-124a-3 was associated with an increased risk of developing metachronous gastric cancers. Assessment of an epigenetic field defect using methylation levels in normal tissue is a promising biomarker for cancer risk that takes account of life history.
Acknowledgments
The authors are grateful to Dr Shigetaka Yoshinaga and Dr Haruhisa Suzuki for patient follow-up and to Eiken for measuring serum pepsinogen levels and serum anti-H. pylori antibody levels of all the patients.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Files in this Data Supplement:
- Data supplement 1 - Online supplement
- Data supplement 2 - Online tables
Footnotes
-
KA and TN contributed equally.
-
Collaborators Shigetaka Yoshinaga; Haruhisa Suzuki.
-
Contributors TU, TN, MF and MI conceptualised the study. KA, TA and SN performed the experiments. TN, NY, TM, CY, IO, TY, MF, TG and MI followed up the patients endoscopically. TS carried out data analysis. TU and KA wrote the manuscript.
-
Funding This study was supported by a grant-in-aid for pioneering basic research from the Ministry of Health, Labour (H20-genome-g-006) and Welfare and by a National Cancer Center Research and Development Fund (H23-A-2), Japan.
-
Competing interests None.
-
Patient consent Written informed consent was obtained from all the patients.
-
Ethics approval Study approved by the institutional review board at each hospital.
-
Provenance and peer review Not commissioned; externally peer reviewed.