Article Text
Abstract
Objective Patients with primary sclerosing cholangitis (PSC) were previously shown to display a bacterial gut dysbiosis but fungal microbiota has never been examined in these patients. The aim of this study was to assess the fungal gut microbiota in patients with PSC.
Design We analysed the faecal microbiota of patients with PSC and concomitant IBD (n=27), patients with PSC and no IBD (n=22), patients with IBD and no PSC (n=33) and healthy subjects (n=30). Bacterial and fungal composition of the faecal microbiota was determined using 16S and ITS2 sequencing, respectively.
Results We found that patients with PSC harboured bacterial dysbiosis characterised by a decreased biodiversity, an altered composition and a decreased correlation network density. These alterations of the microbiota were associated with PSC, independently of IBD status. For the first time, we showed that patients with PSC displayed a fungal gut dysbiosis, characterised by a relative increase in biodiversity and an altered composition. Notably, we observed an increased proportion of Exophiala and a decreased proportion of Saccharomyces cerevisiae. Compared with patients with IBD and healthy subjects, the gut microbiota of patients with PSC exhibited a strong disruption in bacteria-fungi correlation network, suggesting an alteration in the interkingdom crosstalk.
Conclusion This study demonstrates that bacteria and fungi contribute to gut dysbiosis in PSC.
- gut microbiota
- fungi
- inflammatory bowel disease
- primary sclerosing cholangitis
Statistics from Altmetric.com
Significance of this study
What is already known on this subject?
Bacterial gut dysbiosis has been shown to occur in patients with primary sclerosing cholangitis (PSC).
PSC is frequently associated with IBD.
Patients with IBD display both bacterial and fungal gut dysbiosis.
What are the new findings?
Patients with PSC bear fungal gut dysbiosis, combining an altered composition and increased biodiversity.
The signature of fungal gut dysbiosis in patients with PSC is distinct from that of patients with IBD.
In patients with PSC, the gut microbiota is characterised by a disruption in the correlation network between bacteria and fungi.
How might it impact on clinical practice in the foreseeable future?
These data suggest that gut fungi could contribute to the pathogenesis of PSC and therefore should be considered as a new target for future therapeutic strategies.
Introduction
Primary sclerosing cholangitis (PSC) is a chronic cholestatic liver disease characterised by inflammatory and fibro-obliterative lesions of the biliary tract. Disease course in PSC is highly variable, but often progressive, leading to cirrhosis. Effective medical therapy is currently lacking so that liver transplantation is the only life-extending therapeutic intervention in patients with end-stage PSC liver disease.1 2 Noticeably, a majority (up to 80%) of subjects with PSC have a concomitant IBD, most often UC.3 Both IBD and PSC are multifactorial diseases the pathogenesis of which remains to elucidate. They share a number of common genetic risk factors,4 although genetic factors account for only a small fraction of disease liability in PSC.5 6 There is evidence to indicate that the gut microbiota is altered and triggers an exacerbated immune response in IBD.7 Alterations in the composition of the gut microbiota that were found in IBD include a reduction of microbial diversity, a decrease in some species of the Firmicutes phylum and an increase in species of the Proteobacteria phylum.8 9 Recently, several studies have shown that the composition of the gut microbiota was altered also in patients with PSC.10–19 Alterations that have been reported in PSC consist in a reduction of biodiversity, a decrease in bacteria of the Firmicutes phylum (such as Faecalibacterium) as well as an increase in some Proteobacteria (such as Morganella) and in some bacteria usually found in the oral microbiota (such as Veillonella).11 13–16 18 All studies in PSC have been focused on bacterial dysbiosis. However, other types of microorganisms colonise the human gut including fungi. Fungal dysbiosis has been described in alcoholic liver disease and cirrhosis,20 21 as well as in IBD.22 In the latter case, biodiversity was decreased and the fungi-to-bacteria ratio increased, notably in patients with IBD flare.22 So far, no study has addressed potential changes in fungal microbiota (mycobiota) in PSC. Yet, a number of observations point out to a possible role of fungi in the pathogenesis or progression of PSC. First, anti-Saccharomyces cerevisiae antibodies are highly prevalent in patients with PSC.23 Second, bile can be colonised by Candida albicans in patients with PSC and the presence of Candida in bile was shown to be associated with a poor prognosis in these patients.24 Third, genetic variants of caspase recruitment domain-9, a protein involved in the innate immunity against fungi, provide susceptibility to both PSC and IBD.4 Therefore, in keeping with these lines, we hypothesised that the fungal microbiota might be altered in patients with PSC. To investigate this hypothesis, we analysed the fungal microbiota in a cohort of patients with PSC, including patients with and without IBD, in comparison with patients with IBD alone and with healthy subjects.
Study population and methods
Study population
Patients with PSC and/or IBD were recruited in the Hepatology and Gastroenterology Departments of Saint-Antoine Hospital, Paris, France. The diagnosis of PSC, including small-duct PSC, was established according to the guidelines of the European Association for the Study of the Liver.25 All patients with PSC included in the study underwent screening for IBD. IBD diagnosis was established according to clinical, radiological, endoscopic and histological criteria and only patients in remission were included. Remission status was attested by normal C-reactive protein concentration in serum and the absence of clinical symptoms, based on expert clinicians’ assessment. Exclusion criteria were the following: less than 18 years of age, secondary sclerosing cholangitis, decompensated cirrhosis, previous liver transplantation, active IBD or previous colon surgery, and intake of antibiotics or of colon-cleansing preparation within the last 3 months preceding stool collection. Healthy subjects were recruited via the healthy donor biobank of the Gastroenterology Department in Saint-Antoine Hospital. All recruited patients and healthy subjects provided written informed consent before stool donation.
Collection of clinical data
Clinical data were collected by reviewing medical charts. The following data were recorded: the date of PSC diagnosis, the history of acute cholangitis and endoscopic treatment, the date of IBD diagnosis, IBD topography and medications for PSC or IBD. In patients with PSC, serum liver tests were performed at the time of stool collection and included alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), gamma-glutamyl transpeptidase (GGT), total serum bilirubin and serum albumin. In these patients, liver stiffness assessed by transient elastography at the time of stool collection ±12 months was recorded when available. Cirrhosis was defined histologically or by liver stiffness >14.4 kPa.26
Stool collection and DNA extraction
Whole stools were collected in sterile boxes, immediately homogenised, and aliquots of 0.2 g were frozen at −80°C for further analysis. DNA was extracted from the faecal samples, as described.22 Following microbial lysis by both mechanical and chemical methods, nucleic acids were precipitated in isopropanol for 10 min at room temperature, incubated for 15 min on ice and centrifuged for 30 min at 20 000 g and 4°C. Pellets were suspended in 450 µL of phosphate buffer and 50 µL of potassium acetate. After RNase treatment and DNA precipitation, nucleic acids were recovered via centrifugation at 20 000 g and 4°C for 30 min. The DNA pellet was suspended in 80 mL of trypsin-EDTA buffer. DNA samples were then subjected to 16S and ITS2 sequencing for bacterial and fungal microbiota analyses, respectively.
16S and ITS2 DNA sequencing
Bacterial diversity in stools was determined by targeting a portion of the ribosomal genes in extracted DNA. A 16S DNA gene fragment comprising the V3 and V4 hypervariable regions (16S sense 5′-TACGGRAGGCAGCAG-3′ and antisense 5′-CTACCNGGGTATCTAAT-3′) was amplified using an optimised and standardised 16S amplicon library preparation protocol (Metabiote, GenoScreen). Briefly, 16S DNA PCR was performed using 5 ng of genomic DNA according to the manufacturer’s protocol (Metabiote), 192 bar-coded primers (Metabiote MiSeq Primers) at final concentrations of 0.2 μmol/L and an annealing temperature of 50°C for 30 cycles. The PCR products were purified using an Agencourt AMPure XP-PCR purification system (Beckman Coulter, Brea, CA, USA), quantified according to the manufacturer’s protocol and multiplexed at equal concentrations. Sequencing was performed using a 300 bp paired-end sequencing protocol on an Illumina MiSeq platform (Illumina, San Diego, CA, USA) at GenoScreen, Lille, France. Raw paired-end reads were subjected to the following processes: (1) quality filtering using the PRINSEQ-lite PERL script,27 by truncating the bases from the 3′ end, that did not exhibit a quality <30, based on the Phred algorithm; (2) paired-end read assembly using fast length adjustment of short reads to improve genome assemblies,28 with a minimum overlap of 30 bases and a 97% overlap identity; and (3) searching for and removing both forward and reverse primer sequences using CutAdapt, with no mismatches allowed in the primer sequences. Assembled sequences, for which perfect forward and reverse primers were not found, were eliminated. A similar approach was used for fungi microbiota using the primers ITS2 (sense 5′-GTGARTCATCGAATCTTT-3′ and antisense 5′-GATATGCTTAAGTTCAGCGGGT-3′) and the optimised and standardised ITS2 amplicon library preparation protocol (Metabiote, GenoScreen).
16S and ITS2 sequence analyses
The sequences were demultiplexed and quality filtered using the QIIME V.1.9.1 software package.29 The sequences were then assigned to operational taxonomic units (OTU) using the UCLUST algorithm with a 97% pairwise identity threshold and classified taxonomically using the Silva reference database (version 132) for bacteria and the UNITE ITS database (alpha version 12_11) for fungi.30 Rarefaction was performed (11 000 and 2900 sequences per sample for 16S and ITS2, respectively) and used to compare the relative abundance of OTUs across samples. Alpha diversity was estimated by the Shannon diversity index or the number of observed species. Beta diversity was measured by a Bray-Curtis distance matrix and was used to build principal coordinate analysis plots. Raw sequence data are accessible in the European Nucleotide Archive (accession number PRJEB30862).
Statistical analyses
Patients’ characteristics were expressed as median (range) and compared using Mann-Whitney or Χ2 tests as appropriate. GraphPad Prism V.6.0 (San Diego, CA, USA) was used for analyses and graph preparation. Statistical analyses were performed using the two-tailed non-parametric Mann-Whitney test or Kruskal-Wallis test with Dunn’s multiple comparison test. Statistical significance of sample grouping for beta diversity was performed using analysis of similarity method (9999 permutations). Multivariate association test with linear models (MaAsLin), a multivariate statistical framework, was used to find associations between clinical metadata and microbial community abundance using default setting (p values <0.05 with q values <0.25 were considered significant).31 Correlation networks were built using Spearman’s correlation or distance correlation tests,22 and figures were built using Cytoscape V.3.6.0. The p values were corrected using the Benjamini and Hochberg procedure to control for the false discovery rate. The relative connectedness of the networks was calculated as the ratio between the number of significant interactions (edges) and the number of taxa (nodes) in the network. In all statistical analyses, differences with p values <0.05 were considered significant.
Results
Characteristics of the study population
The study population included four groups: patients with PSC and no IBD (PSC_only, n=22), patients with PSC and IBD (PSC_IBD, n=27), patients with IBD and no PSC (IBD_only, n=33) and healthy subjects (n=30). To limit the potential effects of active intestinal inflammation on gut microbiota, all patients with IBD included in the present study were in remission. Table 1 shows the study population’s characteristics. Patients with PSC are presented together and separately depending on whether they had or not concomitant IBD. A minority of patients with PSC had cirrhosis (approximately 10%) and none of them had decompensated cirrhosis. A large majority of patients with PSC (more than 90%) were treated with ursodeoxycholic acid (UDCA). The two groups of patients with PSC, that is, PSC_only and PSC_IBD, were comparable, notably in terms of age, gender, body mass index, UDCA treatment, PSC duration and severity, notably assessed by the Mayo risk score,32 liver stiffness and the presence or not of cirrhosis. The only significant difference was the prevalence of overlap syndrome (PSC associated with autoimmune hepatitis), which was diagnosed in four patients without IBD versus none of the patients with IBD. Likewise, the two groups of patients with IBD, that is, PSC_IBD and IBD_only, were comparable in terms of age, body mass index, IBD duration and phenotype, as well as 5-aminosalicylic acid and steroid treatments. However, patients with PSC_IBD were more frequently males and had more often extensive colitis (Montreal E3) than patients with IBD_only.
Study population’s characteristics
Bacterial dysbiosis in PSC
The bacterial fraction of the gut microbiota was analysed using 16S sequencing. The analysis of beta diversity showed a clustering of samples according to the study group (figure 1A; online supplementary figure 1A,B). The difference observed in beta diversity was statistically significant between patients with IBD either alone or with PSC on the one hand and healthy subjects on the other. The difference between patients with PSC_IBD and those with IBD_only was also statistically significant, whereas the difference between patients with PSC_only and healthy subjects showed a trend towards significance (p=0.09). These results indicated that both IBD and PSC were linked to intestinal dysbiosis. Alpha diversity, assessed by the Shannon and Chao1 indexes, was not significantly different between patients with IBD_only and healthy subjects, which could be explained by the fact that all patients with IBD were in remission. Yet, patients with PSC_IBD showed significantly lower alpha diversity than either patients with IBD_only or healthy subjects (figure 1B,C). This suggested that alterations in the gut bacterial microbiota diversity were aggravated when PSC was superimposed on IBD. In keeping with previous work, we found that in all study groups, the bacterial gut microbiota was dominated by bacteria from the Firmicutes, Bacteroidetes, Actinobacteria and Proteobacteria phyla (figure 2A) and from the Ruminococcaceae, Lachnospiraceae, Veillonellaceae, Rikenellaceae, Bifidobacteriaceae and Enterobacteriaceae families (figure 2B). We sought to determine if bacterial features were associated with disease phenotype, using a MaAsLin, to over-ride the effect of potential confounding factors such as age, gender, smoking or treatment. Associations between clinical features and microbiota bacteria thus found are shown in online Supplementary file 2. This analysis showed that the PSC status was associated with alterations in the abundance of several taxa, notably a decrease in members of the Firmicutes phylum, except for Veillonella the abundance of which was increased, and an increase in members of the Proteobacteria phylum (figure 2C; online Supplementary file 2). We also found negative correlations between the abundance of several bacteria OTUs (including the Alistipes, Blautia, Coprococcus and Roseburia genera and the Eubacterium hallii species) and serum liver tests (notably total bilirubinaemia) in patients with PSC (figure 2D). Furthermore, treatment with UDCA was found to be negatively associated with bacteria from the Sphingomonadaceae family (online Supplementary file 2). Bacterial taxa found to be associated with the IBD status are shown in online Supplementary file 3A.
Supplementary file 1
Supplementary file 2
Supplementary file 3
Biodiversity of the bacterial gut microbiota. The bacterial fraction of gut microbiota was analysed using 16S sequencing in healthy subjects and in patients with IBD_only, primary sclerosing cholangitis (PSC)_IBD or PSC_only. (A) Beta diversity was assessed by principal coordinate analysis of Bray-Curtis distance. PC1, PC2 and PC3 represent the top three principal coordinates that captured most of the diversity. The fraction of diversity captured by the coordinate is given as a percentage. Each sample is coloured according to the study group; analysis of similarity (ANOSIM) method was used for comparisons. Alpha diversity was estimated by (B) Shannon and (C) Chao1 indexes; Kruskal-Wallis test with Dunn’s multiple comparison test were used for comparisons. Means and SEM are indicated in panels (B) and (C). In all panels, *p<0.05; **p<0.01; ***p<0.001.
Composition of the bacterial gut microbiota. Global composition of bacterial microbiota at (A) the phylum and (B) family levels was determined in the four study groups (as defined in figure 1), and expressed as the relative abundance of operational taxonomic units (OTU) in each group. (C) Bacterial taxa associated with primary sclerosing cholangitis (PSC) were identified using multivariate association test with linear models (MaAsLin) multivariate analysis. For each taxon, the fold change was calculated as the ratio of mean abundance between PSC cases and healthy subjects. (D) Correlations between bacterial OTUs and liver tests in patients with PSC were analysed. Only OTUs present in >50% of the samples were taken into account, and only significant correlations (p<0.05 after correction for the false discovery rate with Benjamini and Hochberg procedure) are shown. ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, gamma-glutamyl transpeptidase; ULN, upper limit of normal.
Altered bacterial correlation network in PSC
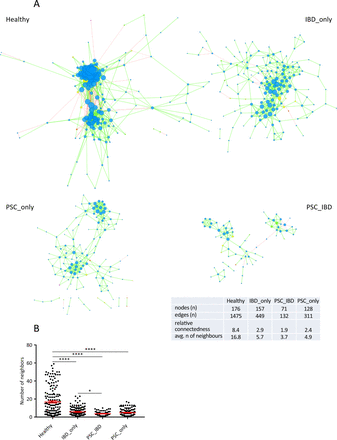
Next, we built a bacterial abundance correlation network to evaluate the bacterial ecosystem structure. The detailed correlations between bacteria are reported in online Supplementary file 4. A rich and complex network of correlations between bacteria was observed in healthy controls (figure 3A, left upper panel). In comparison, the density of bacterial correlation network was decreased in patients with IBD_only and to an even greater extent, in patients with PSC, combined or not with IBD, as attested by a decreased relative connectedness and a lower number of neighbours (figure 3A,B). There was no statistically significant difference between PSC_only and PSC_IBD groups for these two parameters, but the network in PSC_IBD was smaller (nodes n=71) than in PSC_only (nodes n=128), suggesting a deleterious additive effect of PSC and IBD.
Supplementary file 4
Correlation networks of bacterial gut microbiota. Correlation networks between bacteria were built using Spearman correlation test in the four study groups (as defined in figure 1). (A) Abundance correlation networks are shown, in which each circle (node) represents an operational taxonomic unit (OTU), its colour, the bacterial phylum (blue, Firmicutes; green, Bacteroidetes; yellow, Actinobacteria; orange, Proteobacteria), and its size, the number of direct edges. Edges indicate the magnitude of distance correlation (positive in green, negative in red). Only OTUs present in >50% of the samples were taken into account, and only significant correlations (p<0.05 after correction for false discovery rate using Benjamini and Hochberg procedure) are shown. The table in the inset shows the networks' parameters. The relative connectedness of the networks was calculated as the ratio between the number of significant interactions (edges) and the number of taxa (nodes) in the network. (B) Number of neighbours; means and SEM are indicated. *p<0.05; ****p<0.0001. avg, average; PSC, primary sclerosing cholangitis.
Fungal dysbiosis in PSC
Using ITS2 sequencing, we then assessed the composition of the fungal gut microbiota in the study population. The analysis of beta diversity showed a clustering of samples according to the study group (figure 4A; online Supplementary file 5A,B). Differences were statistically significant between healthy subjects and patients with either IBD_only or PSC_only, as well as between patients with PSC_IBD and patients with either IBD_only or PSC_only. The analysis of alpha diversity, assessed by the Shannon and Chao1 indexes, showed no significant difference between the four study groups (figure 4B; online Supplementary file 5C). However, globally taken, patients with PSC (ie, PSC_only combined with PSC_IBD) displayed a significantly higher fungal diversity assessed by the Shannon index than patients with IBD_only and a trend (p=0.08) towards higher diversity than healthy subjects (figure 4C; online Supplementary file 5D). In all groups, the fungal microbiota was dominated by two phyla: Ascomycota and Basidiomycota (figure 4D). The most dominant genera identified were: Saccharomyces, Debaryomyces, Candida, Penicillium, Fusarium, Kluyveromyces, Exophiala, Alternaria and Filobasidium (figure 4E). We then used MaAsLin to identify the fungi taxa potentially associated with the disease phenotype. After adjusting for confounding factors, we found that PSC was associated with increases in Exophiala genus and Sordariomycetes class (figure 4F). The increased abundance of Exophiala was particularly striking even if it was observed in only a few patients with PSC (figure 4F; online Supplementary file 5E). Conversely, PSC was associated with a decrease in the Saccharomycetales order, Saccharomycetes class, Saccharomycetaceae family and S. cerevisiae species (figure 4F; online Supplementary file 5F). Noticeably, we did not observe strong associations between IBD status and fungi taxa. Only minor changes in Saccharomyces abundance were observed (online Supplementary file 3B). These results are different from those we previously reported in a large cohort of patients with IBD. The difference is explained by the absence of patients with active IBD in the present study, while in our previous study, we found that changes in fungi microbiota were mostly driven by active inflammation.22
Supplementary file 5
Biodiversity and composition of the fungal gut microbiota. The fungal fraction of gut microbiota was analysed using ITS2 sequencing in the four study groups (as defined in figure 1). (A) Beta diversity was assessed by principal coordinate analysis of Bray-Curtis distance. PC1, PC2 and PC3 represent the top three principal coordinates that captured most of the diversity. The fraction of diversity captured by the coordinate is given as a percentage. Each sample is coloured according to the study group; analysis of similarity (ANOSIM) method was used for comparisons. Alpha diversity was estimated by the Shannon index (B) in the four study groups and (C) in all patients with primary sclerosing cholangitis (PSC) (with or without IBD) in comparison with healthy subjects and patients with IBD_only; Kruskal-Wallis test with Dunn’s multiple comparison test were used for comparisons. Global composition of fungal microbiota at (D) the phylum and (E) genus levels was determined in the four study groups, and expressed as the relative abundance of operational taxonomic units (OTU) in each group. (F) Fungal taxa associated with PSC were identified using multivariate association test with linear models (MaAsLin) multivariate analysis. For each taxon, the fold change was calculated as the ratio of mean abundance between PSC cases and healthy subjects. Means and SEM are indicated in panels (B) and (C). In all panels, *p<0.05.
Altered interkingdom network in PSC
To explore the equilibrium between fungi and bacteria diversity in the gut, we determined the fungi-to-bacteria diversity ratio. This ratio was significantly higher in patients with PSC_IBD than in those with IBD_only or in healthy subjects (figure 5A; online Supplementary file 6A). Likewise, when considered globally, patients with PSC had a higher ITS2/16S diversity ratio than patients with IBD_only (figure 5B; online Supplementary file 6B). No significant difference of the fungi-to-bacteria diversity ratio was observed between healthy subjects and patients with PSC_only (p=0.23 for Shannon) or all patients with PSC (p=0.11 for Shannon).
Supplementary file 6
Fungi-bacteria networks. To address the equilibrium between gut bacteria and fungi in the four study groups (as defined in figure 1), ITS2/16S diversity ratio was estimated by the Shannon index (A) in the four study groups and (B) in all patients with primary sclerosing cholangitis (PSC) (with or without IBD) in comparison with healthy subjects and patients with IBD_only; Kruskal-Wallis test with Dunn’s multiple comparison test were used for comparisons. (C) Correlation networks at the family and genus levels were built in the four groups using distance correlation test. Abundance correlation networks are shown, in which each node represents a family or a genus, its shape, the kingdom (square, fungi; circle, bacteria), its colour, the phylum (pink, Ascomycota; purple, Basidiomycota; blue, Firmicutes; green: Bacteroidetes; yellow, Actinobacteria; orange, Proteobacteria), and its size, the number of direct edges. Edges indicate the magnitude of distance correlation (positive in green, negative in red, according to Spearman test). Only family and genus present in >20% of the samples were taken into account, and only significant correlations (p<0.05 after correction for false discovery rate using Benjamini and Hochberg procedure) are shown. The table in the inset shows the networks' parameters. The relative connectedness of the networks was calculated as the ratio between the number of significant interactions (edges) and the number of taxa (nodes) in the network. (B) Number of neighbours. Means and SEM are indicated in panels (A), (B) and (D). In all panels, *p<0.05; **p<0.01. avg, average.
Next, we asked whether bacterial and fungi microbiota interactions were altered according to the disease phenotype. To address this question, we built an abundance correlation network at the family and genus level according to the study group. Detailed correlations between bacteria and fungi in each group are reported in online Supplementary file 7. The density of fungi-bacteria network was unchanged in patients with IBD_only, in comparison with healthy subjects (figure 5C,D). In PSC_only and PSC_IBD groups, the interkingdom network was disrupted, as illustrated by a decrease in the number of nodes and edges compared with the healthy or IBD_only groups (figure 5C,D; online Supplementary file 8). Indeed, the number of connected nodes in the largest connected network was lower in patients with PSC than in the other groups (50, 19, 11 and 8 nodes in healthy subjects, IBD_only, PSC_IBD and PSC_only, respectively). Moreover, in PSC_only, the fungi-bacteria network appeared to be even scarcer than in PSC_IBD, with a significantly lower number of neighbours (figure 5D). These results indicated that PSC was associated with an alteration of the bacterial-fungi interactions.
Supplementary file 7
Supplementary file 8
Discussion
In this study, we showed that patients with PSC bore a bacterial gut dysbiosis and a fungal gut dysbiosis, compared with healthy subjects and patients with IBD_only. A bacterial faecal dysbiosis was previously reported in patients with PSC.10–19 Besides confirmation, we herein provide new data, showing that bacterial microbiota is characterised by alterations in its biodiversity and composition in PSC. In keeping with previous reports,13 14 16 19 we found that PSC was associated with a specific bacterial microbiota signature, distinct from that associated with IBD. Our present results confirm previous findings regarding the altered composition of bacterial microbiota in patients with PSC such as the decreased proportion of Faecalibacterium and of Ruminococcus genera,14 and the increased proportion of Veillonella.11 13–16 18 The consistently increased proportion of Veillonella found in several studies could suggest that Veillonella is involved in PSC pathogenesis. However, the abundance of Veillonella is not specifically related to PSC and is more likely a marker of advanced liver disease.13 33 34 We found a negative correlation between Blautia abundance and serum bilirubin, as well as a decrease in the abundance of Blautia in the microbiota of patients with PSC, whereas Torres et al previously reported an increase.12 In this latter study, Torres et al used mucosal biopsies obtained during colonoscopy to analyse the gut microbiota, which can explain the discrepancy with our findings because of the drastic effects of colon-cleansing products administered before colonoscopy on gut microbiota. Indeed, more recently, Torres et al analysed the gut microbiota from stool samples instead of biopsies and like ourselves, found a decreased abundance of Blautia in patients with PSC.19
We also found correlations between several bacteria of the gut microbiota and liver tests in patients with PSC. The abundance of Alistipes was negatively correlated with the serum concentrations of ALT, ALP and GGT. This bacterium was previously shown to be decreased in the gut microbiota of patients with non-alcoholic fatty liver disease.35 The abundance of E. hallii was also negatively correlated with the serum concentrations of AST, ALP, GGT and total bilirubin. This bacterium is able to produce short-chain fatty acids (butyrate and propionate), which have anti-inflammatory properties.36 E. hallii was previously shown to be decreased in the gut microbiota of patients with IBD,37 and in a murine model of non-alcoholic fatty liver disease.38 It was shown to improve insulin sensitivity in a murine model of diabetes,39 and there is evidence to indicate that this effect could also occur in patients with the metabolic syndrome.40 Most of the correlations we found between gut bacteria and liver tests were with bilirubinaemia, highlighting the strong link between cholestasis and bile acids on the one hand and gut microbiota on the other, as recently reported by Torres et al. 19 It should be emphasised in this respect that bacteria and fungi are able to metabolise bile acids.41 Almost all patients with PSC included in this study were on UDCA treatment (94%), which may have influenced our results. UDCA has been shown to impact the intestinal microbiota in patients with IBD,42 or primary biliary cholangitis,43 but so far, no difference in bacterial gut microbiota has been found in patients with PSC, depending on whether or not they were treated with UDCA.13 19 We herein demonstrated that the network of correlations between gut bacteria was altered with a decreased number of correlations in patients with IBD and to an even greater extent in patients with PSC. These results suggest that the functions of the microbiota acting as a community in the gut may be disturbed in PSC.
For the first time in this study, we report the occurrence of fungal dysbiosis in patients with PSC. PSC-associated fungal dysbiosis was characterised by an increased biodiversity (alpha diversity) compared with healthy subjects or patients with IBD and by an altered composition, including notably a marked increase in the abundance of Exophiala genus in some patients. Exophiala is a fungi genus of the Herpotrichiellaceae family that has been involved in infections known as phaeohyphomycosis, particularly in immunocompromised hosts. Most of these infections affect the skin,44 but Exophiala can also cause systemic infections that involve the lungs or the central nervous system.44 Of particular interest, Oztas et al reported the case of a systemic infection by Exophiala dermatitidis mimicking PSC in a 24-year-old woman without immunodeficiency.45 Other investigators reported the case of an end-stage liver disease caused by E. dermatitidis and characterised by cholestasis and dilatation of intrahepatic bile ducts.46 Combined, these clinical observations and our results suggest that Exophiala could participate in the pathophysiology of PSC, at least in a subgroup of patients, a hypothesis that deserves to be tested in the future. In the microbiota of patients with PSC, we also found a decrease in the proportion of the Saccharomycetaceae family, including the species S. cerevisiae, which has been shown to be decreased in the microbiota of patients with IBD,22 and to have anti-inflammatory properties.22 Future studies should also address the potential implication of S. cerevisiae in the occurrence or progression of PSC. Bacterial and fungal dysbiosis associated with PSC was different from that associated with IBD, which suggests a specificity of dysbiosis in PSC, distinct from that in IBD. The fungal microbiota of patients with PSC displayed an increased biodiversity and its correlation network with bacteria was disrupted, especially in patients with PSC_only. A disrupted correlation network between bacteria and fungi has been previously reported in patients with cirrhosis.21
Our study has some limitations: it is a monocentric study, the population size is relatively small and the design is observational. However, this work has also major strengths including homogeneous populations of patients with stable PSC and quiescent IBD and two groups of similar size for the comparison of patients with PSC, with and without IBD. The fact that patients were recruited in national reference centres for PSC and IBD is a strength as it ensures reliable diagnosis and rigorous phenotype for both diseases. However, patients followed in a reference centre may not be representative of the whole PSC population,1 3 which may also introduce a selection bias. Provided that our results are confirmed by others, one can envision using faecal microbiota-based biomarkers as a diagnostic tool in patients with suspected PSC. This type of strategy has already been proposed,13 16 18 notably by Iwasawa et al who recently reported a characteristic dysbiosis of the salivary microbiota in children with PSC.47 Furthermore, specific alterations of the gut microbiota found in PSC could be used to design new therapeutic strategies, as recently proposed for alcoholic liver disease.48
We conclude from this study that PSC is characterised by an altered fungal gut microbiota associated with an impaired fungi-bacteria correlation network. While their implications remain to be refined, the present findings pave the way for the development of microbiota-based biomarkers and treatment in PSC.
Supplementary file 9
Acknowledgments
The authors acknowledge Yves Chrétien, CRSA, for the formatting of figures.
References
Footnotes
36 CH and HS share senior authorship
SL and AK authors share first authorship
36 ‡The list of members of the Saint-Antoine Inflammatory Bowel Disease (IBD)Network is provided as Supplementary Material.
Contributors SL, AK: study execution, collection and analysis of data, drafting of the manuscript. KBB, MS, SJ: acquisition of data. HS: statistical analyses, interpretation of data, design of the figures. HS, CH: study concept, design and supervision. OC, CH: fund raising for the study. CC, OC, HS, CH: critical revision of the manuscript. All authors approved the final version. Saint-Antoine IBD Network provided the patients.
Funding This study was supported by the Microbiome Foundation.
Competing interests None declared.
Ethics approval The study was approved by the regional ethics committee for human studies (Comité de Protection des Personnes Ile-de-France IV, IRB 00003835 Suivitheque study; registration number 2012/05NICB).
Provenance and peer review Not commissioned; externally peer reviewed.
Collaborators Members of the Saint-Antoine IBD Network: Arrivé Lionel, Beaugerie Laurent, Bourrier Anne, Camus Marine, ChafaiNajim, Chambenois Édouard, ChaputUlriikka, Delattre Charlotte, Martineau Chloé, Cholley Monnier Laurence, DeboveClotilde, Dray Xavier, Fléjou Jean-François, Le Gall Guillaume, Hoyeau Nadia, Kirchgesner Julien, Landman Cecilia, LevèvreJérémie, Marteau Philippe, Nion-Larmurier Isabelle, Ozenne Violaine, Parc Yann,Seksik Philippe, Sokol Harry, Svrcek Magali.
Correction notice This article has been corrected since it published Online First. The collaborator statement has been added.
Patient consent for publication Obtained.